Multiple Imputation and Cross-validation
Introduction
This page contains information of three methods that are implemented
in the psfmi package and that combine Multiple Imputation with Cross-validation for the validation of logistic prediction models. Currently the methods are only available via downloading the
psfmi package via Github. Do this:
install.packages(“devtools”)
library(devtools)
devtools::install_github(“mwheymans/psfmi”)
library(psfmi)
The cross-validation methods are adjustments of the methods described in the paper of Mertens BJ and Miles A.
The methods are implemented in the function psfmi_perform and are called:
cv_MI, cv_MI_RR and MI_cv_naive. An explanation and examples of how to use the
methods can be found below. See also these Vignettes for more explanation of the methods Vignettes.
Examples
- Method cv_MI - Example 1
- Method cv_MI including BW selection - Example 2
- Method cv_MI_RR - Example 1
- Method cv_MI_RR including BW selection - Example 2
- Method MI_cv_naive - Example 1
- Method MI_cv_naive including BW selection - Example 2
Method cv_MI
With this method imputations are implemented as part of the cross-validation procedure. Within each cross-validation fold, imputation is done once. By repeating this process over multiple imputation runs, multiply imputed training and test sets are generated. Model performance is evaluated and tested in the training and test sets respectively. The method can be performed in combination with backward selection in the training set and subsequently testing the performance in the test set. The method can only be performed when the outcome data is complete and the original data, that contains the missing values has to be included.
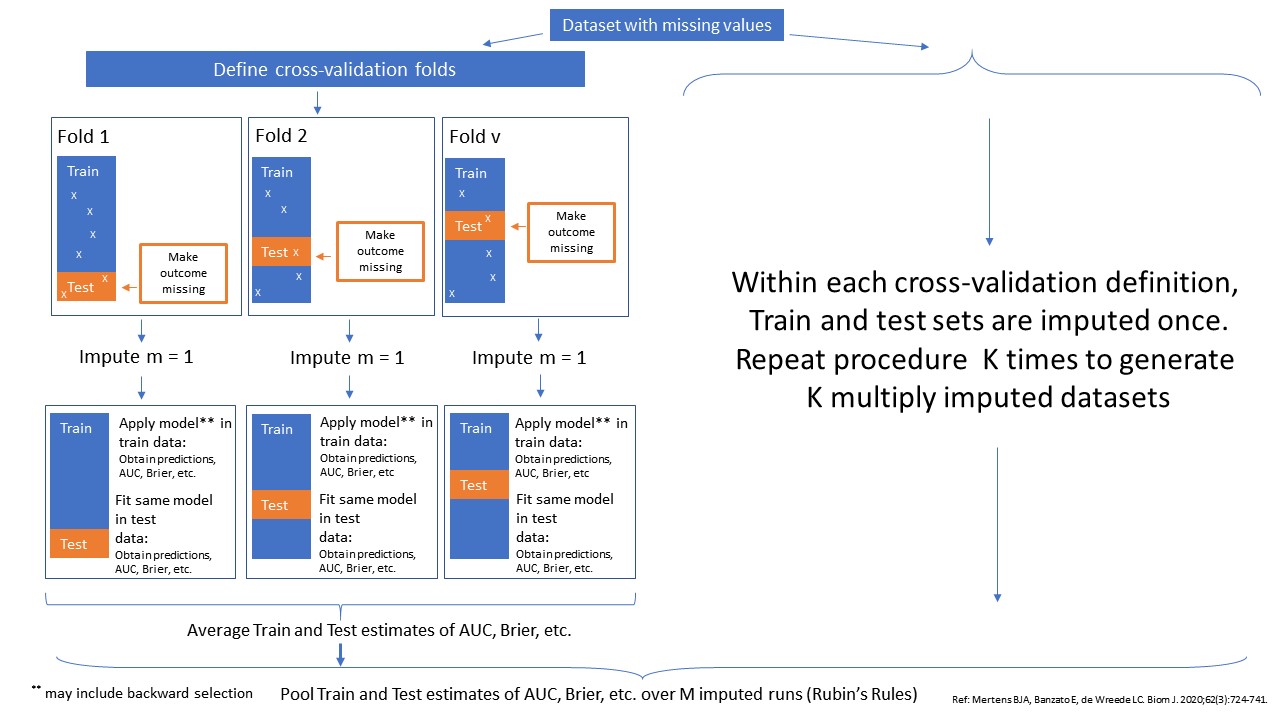
Schematic Overview of Method cv_MI
Back to Methods
Method cv_MI - Example 1
To run the cv_MI method use:
library(psfmi)## Registered S3 methods overwritten by 'car':
## method from
## influence.merMod lme4
## cooks.distance.influence.merMod lme4
## dfbeta.influence.merMod lme4
## dfbetas.influence.merMod lme4pool_lr <- psfmi_lr(data=lbpmilr, formula = Chronic ~ Pain + JobDemands + rcs(Tampascale, 3) +
factor(Satisfaction) + Smoking, p.crit = 1, direction="FW",
nimp=5, impvar="Impnr", method="D1")
res_cv <- psfmi_perform(pool_lr, val_method = "cv_MI", data_orig = lbp_orig, folds=3,
nimp_cv = 2, p.crit=0.2, BW=TRUE, anova_test = "LRT",
miceImp = miceImp, printFlag = FALSE)##
## Imp run 1##
## fold 1## Removed at Step 1 is - Smoking## Removed at Step 2 is - rcs(Tampascale,3)## Removed at Step 3 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 2## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 3## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model##
## Imp run 2##
## fold 1## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 2## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 3## Removed at Step 1 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the modelres_cv## $pool_stats
## Train Test
## AUC 0.8971709 0.8433000
## Scaled Brier 0.4871927 0.3341000
## R2 0.5892451 0.4623542
##
## $LP_val
## (Intercept) lp_test
## 0.06984537 0.75739579
##
## $auc_test
## 95% Low AUC 95% Up
## AUC (logit) 0.7634 0.8433 0.8998Back to Examples
Method cv_MI including BW selection - Example 2
To run the cv_MI method including BW selection use:
library(psfmi)
pool_lr <- psfmi_lr(data=lbpmilr, formula = Chronic ~ Pain + JobDemands + rcs(Tampascale, 3) +
factor(Satisfaction) + Smoking, p.crit = 1, direction="FW",
nimp=5, impvar="Impnr", method="D1")
res_cv <- psfmi_perform(pool_lr, val_method = "cv_MI", data_orig = lbp_orig, folds=3,
nimp_cv = 2, p.crit=0.2, BW=TRUE, anova_test = "LRT",
miceImp = miceImp, printFlag = FALSE)##
## Imp run 1##
## fold 1## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 2## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 3## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## Imp run 2##
## fold 1## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 2## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 3## Removed at Step 1 is - Smoking## Removed at Step 2 is - rcs(Tampascale,3)## Removed at Step 3 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the modelres_cv## $pool_stats
## Train Test
## AUC 0.8942859 0.8454000
## Scaled Brier 0.4681196 0.3375954
## R2 0.5719065 0.4746996
##
## $LP_val
## (Intercept) lp_test
## 0.03880662 0.84650684
##
## $auc_test
## 95% Low AUC 95% Up
## AUC (logit) 0.7416 0.8454 0.9124Back to Examples
Method cv_MI_RR
The method cv_MI_RR uses multiple imputation within the cross-validation definition. The pooled model is analyzed in the training data and subsequently tested in the test data. The method can be performed in combination with backward selection of the pooled model in the training set and subsequently testing the performance of the pooled model in the test set. The method can only be performed when the outcome data is complete.
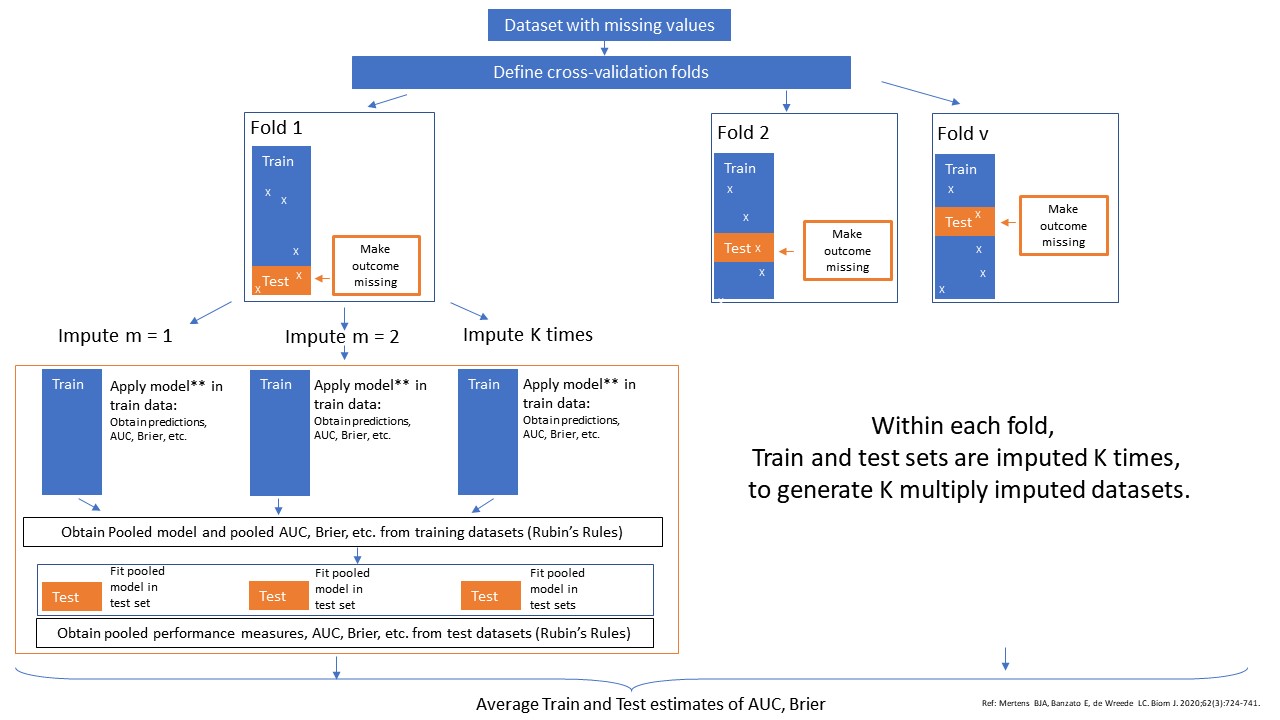
Schematic Overview of Method cv_MI_RR.
Back to Methods
Method cv_MI_RR - Example 1
To run the cv_MI method use:
library(psfmi)
pool_lr <- psfmi_lr(data=lbpmilr, formula = Chronic ~ Pain + JobDemands + rcs(Tampascale, 3) +
factor(Satisfaction) + Smoking, p.crit = 1, direction="FW",
nimp=5, impvar="Impnr", method="D1")
res_cv <- psfmi_perform(pool_lr, val_method = "cv_MI_RR", data_orig = lbp_orig,
folds = 4, nimp_mice = 5, p.crit=0.2, BW=TRUE,
miceImp = miceImp, printFlag = FALSE)##
## fold 1## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 2## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 3## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 4## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the modelres_cv## $stats
## Train Test
## AUC 0.8961433 0.8562267
## Brier scaled 0.4651623 0.3008909
## Rsq 0.5776141 0.5816360
##
## $slope
## Intercept Slope
## 0.07361578 0.84008113Back to Examples
Method cv_MI_RR including BW selection - Example 2
To run the cv_MI_RR method including backward selection:
library(psfmi)
pool_lr <- psfmi_lr(data=lbpmilr, formula = Chronic ~ Pain + JobDemands + rcs(Tampascale, 3) +
factor(Satisfaction) + Smoking, p.crit = 1, direction="FW",
nimp=5, impvar="Impnr", method="D1")
res_cv <- psfmi_perform(pool_lr, val_method = "cv_MI_RR", data_orig = lbp_orig,
folds = 4, nimp_mice = 5, p.crit=0.2, BW=TRUE,
miceImp = miceImp, printFlag = FALSE)##
## fold 1## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 2## Removed at Step 1 is - Smoking## Removed at Step 2 is - rcs(Tampascale,3)## Removed at Step 3 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 3## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands##
## Selection correctly terminated,
## No more variables removed from the model##
## fold 4## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the modelres_cv## $stats
## Train Test
## AUC 0.8864668 0.8380625
## Brier scaled 0.4635775 0.3267663
## Rsq 0.5611362 0.4378292
##
## $slope
## Intercept Slope
## -0.1363122 0.7899024Back to Examples
Method MI_cv_naive
This method applies cross-validation after Multiple Imputation. The same folds are used in each multiply imputed dataset. Is is possible to do backward selection during cross-validation. How this method works is visualized below.
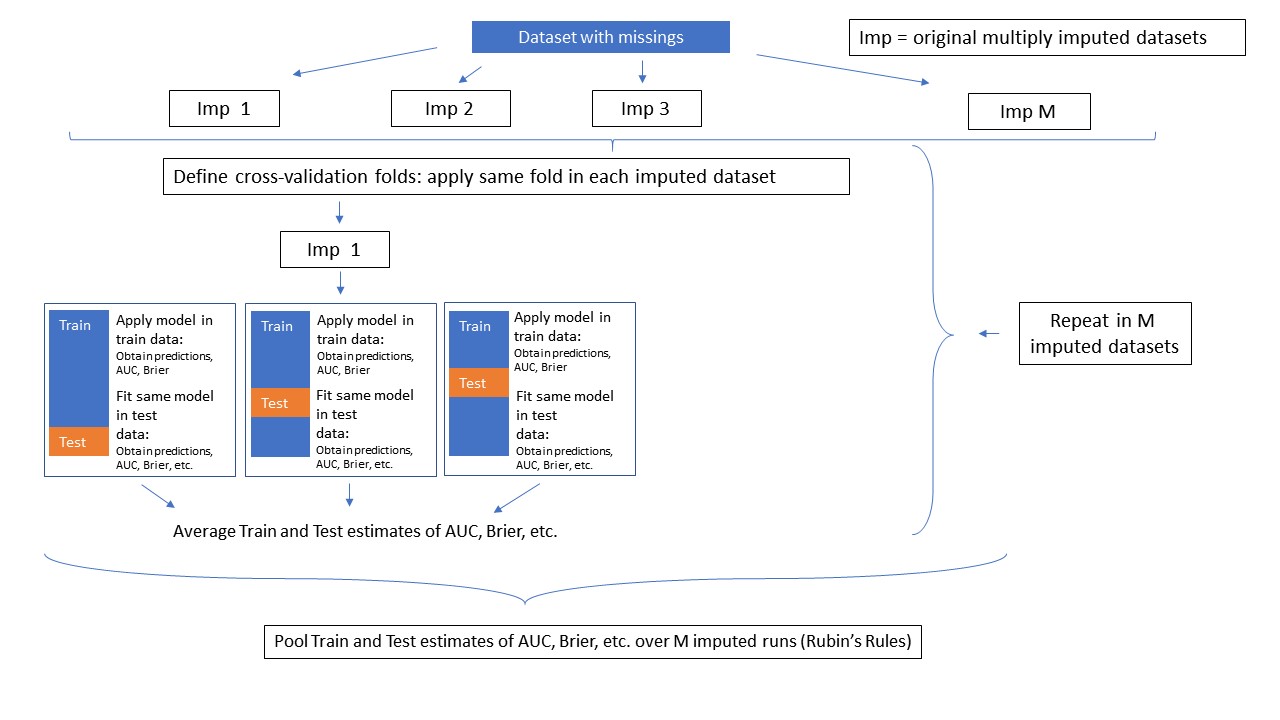
Schematic Overview of Method MI_cv_naive.
Back to Methods
Method MI_cv_naive - Example 1
To run the MI_cv_naive method use:
library(psfmi)
pool_lr <- psfmi_lr(data=lbpmilr, formula = Chronic ~ Pain + JobDemands + rcs(Tampascale, 3) +
factor(Satisfaction) + Smoking, p.crit = 1, direction="FW",
nimp=5, impvar="Impnr", method="D1")
res_cv <- psfmi_perform(pool_lr, val_method = "MI_cv_naive", folds=3, p.crit=1, BW=FALSE)##
## Imputation 1##
## Imputation 2##
## Imputation 3##
## Imputation 4##
## Imputation 5res_cv## $cv_stats
## Train Test
## AUC 0.8920379 0.8410000
## Brier scaled 0.4606837 0.3227124
## R-squared 0.5717383 0.5117027
##
## $auc_test
## 95% Low AUC 95% Up
## AUC (logit) 0.7602 0.841 0.8982
##
## $test_coef
## Intercept Slope
## 0.03362436 0.89480356Back to Examples
Method MI_cv_naive including BW selection - Example 2
To run the MI_cv_naive method by implementing backward variable selection during cross-validation use:
library(psfmi)
pool_lr <- psfmi_lr(data=lbpmilr, formula = Chronic ~ Pain + JobDemands + rcs(Tampascale, 3) +
factor(Satisfaction) + Smoking, p.crit = 1, direction="FW",
nimp=5, impvar="Impnr", method="D1")
res_cv <- psfmi_perform(pool_lr, val_method = "MI_cv_naive", folds=3, p.crit=0.05, BW=TRUE)##
## Imputation 1## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model##
## Imputation 2## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - rcs(Tampascale,3)## Removed at Step 3 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - factor(Satisfaction)## Removed at Step 4 is - Pain##
## Selection correctly terminated,
## No more variables removed from the model##
## Imputation 3## Removed at Step 1 is - Smoking## Removed at Step 2 is - JobDemands## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - Pain##
## Selection correctly terminated,
## No more variables removed from the model##
## Imputation 4## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model##
## Imputation 5## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - rcs(Tampascale,3)## Removed at Step 3 is - Smoking##
## Selection correctly terminated,
## No more variables removed from the model## Removed at Step 1 is - JobDemands## Removed at Step 2 is - Smoking## Removed at Step 3 is - rcs(Tampascale,3)##
## Selection correctly terminated,
## No more variables removed from the modelres_cv## $cv_stats
## Train Test
## AUC 0.8801651 0.8370000
## Brier scaled 0.4477188 0.3724069
## R-squared 0.5402726 0.4857196
##
## $auc_test
## 95% Low AUC 95% Up
## AUC (logit) 0.7479 0.837 0.8989
##
## $test_coef
## Intercept Slope
## -0.03830607 0.98729949Back to Examples
