

Most examples and solutions will be concentrated on Missing Data topics. Posts may also concern Regression modeling, Clinical Prediction models and Spline regression modeling. Most examples originate from FAQs asked during statistical consultations or during courses.
Examples are provided for applied researchers and frequenly in RStudio.
RStudio is an open source free package with a lot of possibilities, making it easy to share code so that you can use the code example and adjust it to your own needs.
With the psfmi_stab function of the psfmi package it is possible to do important stability analyses of the models and predictors selected during backward selection after multiple imputation.

With the psfmi_mm function pooling and selection of (generalized) linear mixed models, i.e linear and logistic mixed models, is possible. With the psfmi_stab function the stability of models after using psfmi_lr, psfmi_coxr and psfmi_mm can be evaluated. The latter function uses (single) bootstrapping for the psfmi_lr and psfmi_coxr functions and cluster bootstrapping for the psfmi_mm function. With the function the bootstrap inclusion frequency of predictors and models can be estimated.

More information about the package can be found on the Cran website.
You can easily install the package by running install.packages("psfmi") in the Console window in Rstudio or R. The development version can be installed from Github by using: install.packages("devtools")
library(devtools)
devtools::install_github("mwheymans/psfmi")
library(psfmi)
If you have questions about the psfmi package send an email to mw.heymans@amsterdamumc.nl or leave a message below. Enjoy using the package!

With the psfmi package it is possible to force variables in the model during backward selection.
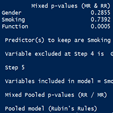
With the psfmi package it is possible to do backward selection for logistic regression models after Multiple Imputation (MI). For backward selection, several variable selection criteria can be used. These criteria are called the D1, D2, D3 and Median P-rule (MPR).

